TOP > Research > Computational Biology Research Core
|
||||
The quantification of complex biological systems requires advanced and innovative modelling techniques. Combining these improved models with experimental methods will allow for detailed analysis of biological systems on all scales beginning from single molecules to entire organs. Biological systems are complex, dynamic and non-linear, which greatly complicates their quantitative analysis, as this requires quantities of data that are too great for current techniques to process. For example, an accurate simulation of protein dynamics requires a time-scale of milliseconds with a time resolution of femtoseconds, resulting in massive amounts of data. This is even more true when simulating molecular networks. Consider the large number of elements that constitute a large-scale system like a cell or organ. Models must not only recreate the behaviour of the individual elements in these networks, they must also reproduce their interactions with one another. Innovations in supercomputing and more efficient algorithms have been instrumental in overcoming these limitations. Taking advantage of these advances, this Core will focus on simulating three phenomena that are key to understanding biological function: 1) milli-second scale simulations of molecular behaviours, 2) simulations of intracellular particles, and 3) simulations of cell behaviour inside a tissue or organ. |
||||
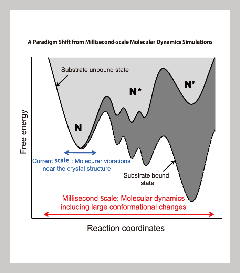
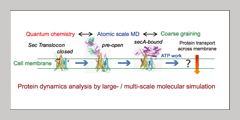
Currently, the best models for proteins come from crystal structure studies. While these offer great detail about the spatial state of the protein, it offers a very small window on the reaction dynamics (see right). Supercomputers and other advance instrumentation will allow us to do multi-scale simulations that span a period of milliseconds, giving us details on previously unseen states. By comparing these predictive models with in vivo experiments, we anticipate revealing greater detail on how proteins respond to their environment over time. The improved resolutions will give us better understanding on how molecular constituents affect enzyme reactions and protein transport, information that can then be applied to more efficient and effective drug design. |
||||
 |
||||
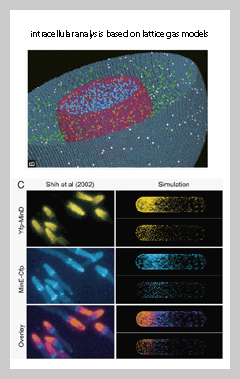
New imaging techniques will allow us to identify previously unclear interplays between macromolecules in situ. Of particular interest will be the investigation of macromolecular crowding, localization, and clustering would affect overall spatio-temporal dynamics a cell. The simulation and analysis techniques we will develop in the core unit will be the basis for prediction and controlling the cell's behavior in response to different stimuli. |
 |
|||
Much like simulating intracellular particles, we aim to create single cell level simulations that describe processes like cellular development, specialization apoptosis that are necessary to realize tissue and organ function. By measuring the biochemical and dynamical cellular interactions, we aim to construct mathematical models that can reproduce these phenomena for the purpose of predicting and controlling tissue behaviour. |
 |
|||